
The knoSYS® system provides labs with a scalable, production-grade informatics infastructure for the analysis, annotation, and interpretation of human next-gen sequence data.
It is a fully integrated system for alignment, variant calling, annotation, multi-sample comparison, analysis, variant classification, report content drafting, and storage of human genome sequence data.
The knoSYS is designed for operational support of research and clinical laboratories using next-generation sequencing technologies and is capable of storing and sharing variants, panels, exomes, and genomes.
 “The disconnect between low-cost sequencing on one hand and high-cost, resource-intensive analysis on the other, can present a barrier to entry for many to the opportunities to apply genomics to urgent medical needs. By partnering with Knome and acquiring a knoSYS system, we have made an important step towards democratizing our pharmacogenomics projects.”
“The disconnect between low-cost sequencing on one hand and high-cost, resource-intensive analysis on the other, can present a barrier to entry for many to the opportunities to apply genomics to urgent medical needs. By partnering with Knome and acquiring a knoSYS system, we have made an important step towards democratizing our pharmacogenomics projects.”
– Corey Nislow, PhD, University of British Columbia
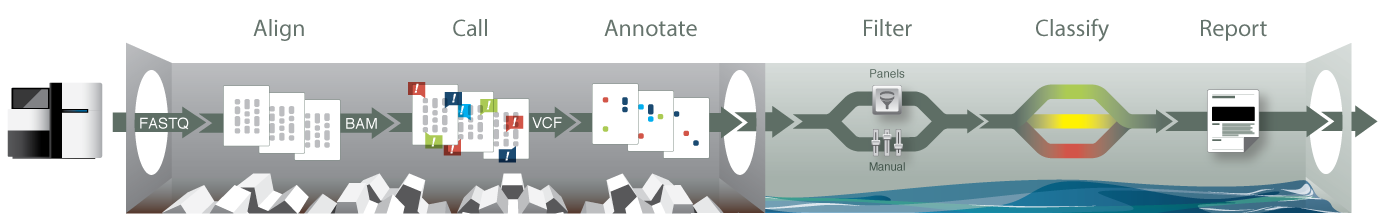
Powerful informatics engine
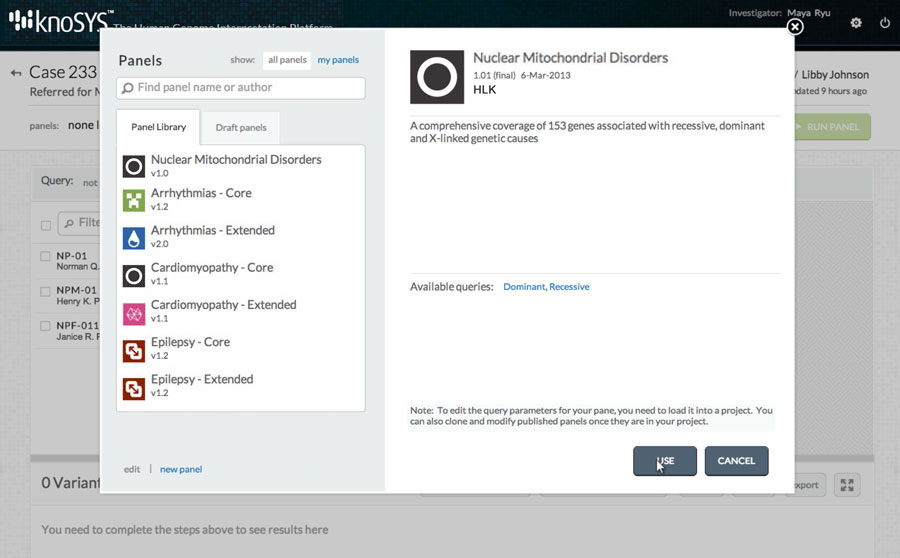
In silico panels encapsulate expertise for efficient variant prioritization
Easy-to-use interface simplifies entire testing process
Bioinformatics is typically a command line driven, highly manual, and ‘finicky’ process. The knoSYS platform provides an easy-to-use graphical user interface that both automates and simplifies the process, reducing the need for dedicated bioinformatics effort to manage the process.
All members of a multidisciplinary lab team can work productively together using the knoSYS. From technologists and bioinformaticians to genetic counselors and lab directors, everyone can access the parts of the NGS analysis process that matter to them.
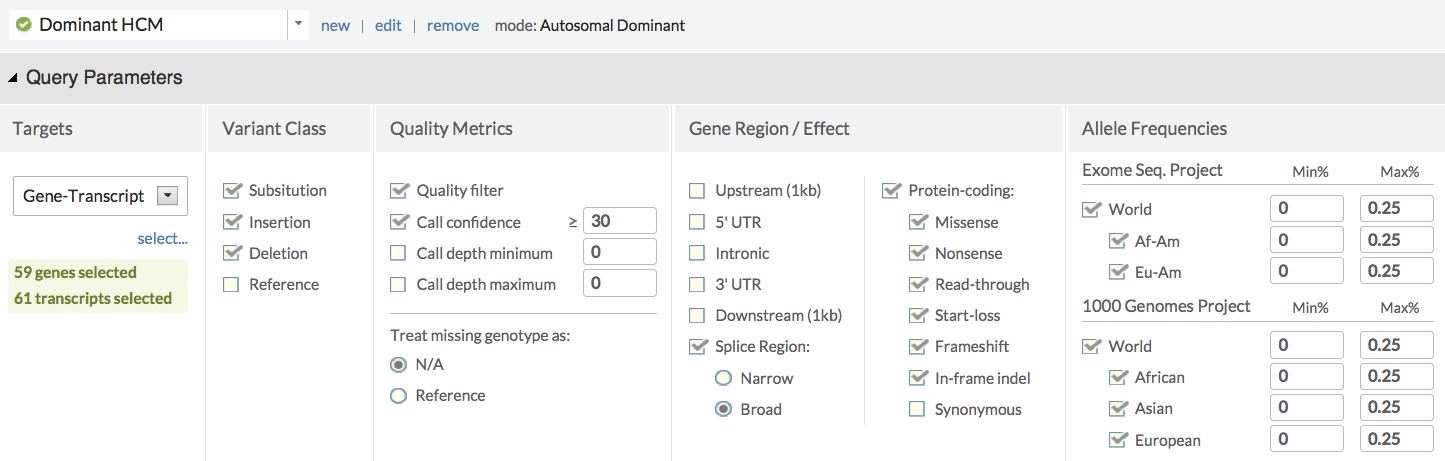
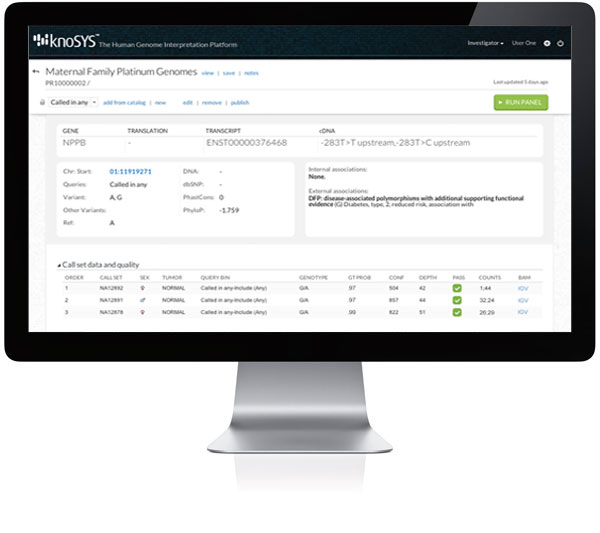
Interactive variant explorer accelerates interpretation and classification

“The knoSYS is an extraordinarily
powerful system. It combines an open
platform for alignment and variant
calling with integrated, user-friendly software for the analysis and
interpretation of gene panels, exomes,
and whole genomes. The knoSYS consistently improves the speed and
quality of our projects, large, andsmall.”
– Massimo Delledonne, PhD, University of Verona
Scalable options to fit every lab
Pay-per-sample and license options are available.
Software only – The knoSYS software can be installed on an organization’s existing, high-performance hardware.
k25 – For labs that focus on targeted panels or moderate number of whole genomes, the k25 hardware is a low-cost entry solution for genomic interpretation.
k100 – The k100 is a high-performance computing cluster with a database server and storage units. And it’s scalable, making it the optimal turnkey informatics solution for high-volume labs.
Hosted – Either hardware model can also be hosted at a secure facility and accessed remotely.